Software Resources
16Stimator
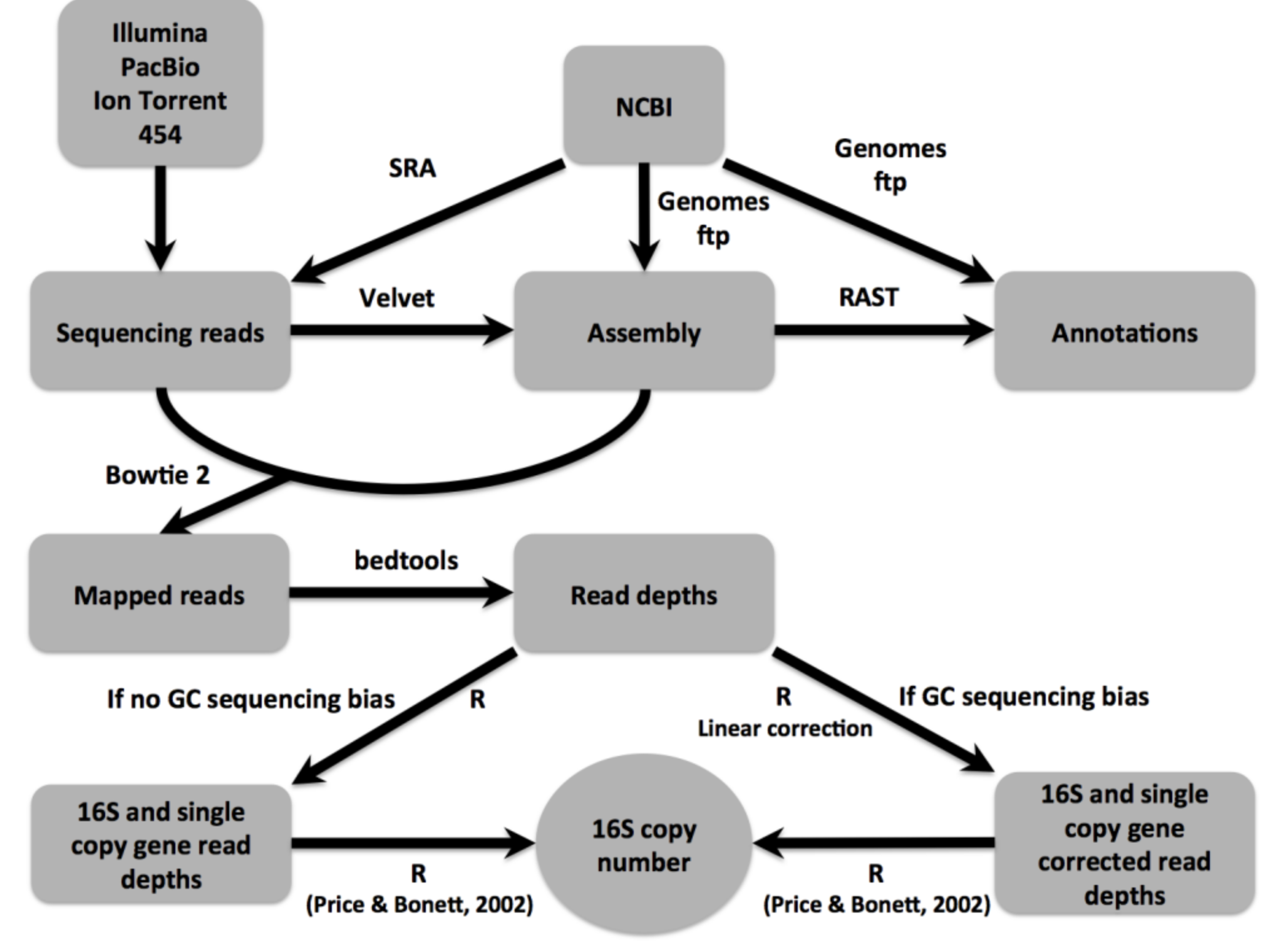
Generate 16S rRNA gene (16S) copy number estimates for bacterial genomes based on comparison of sequencing read depths of ribosomal and single copy gene regions. The method is published in: Perisin et al. 2015. Unfortunately, the publication is not open access (please e-mail to retrieve a copy) but the code of 16Stimator is freely available.
MARTA – Metagenomic and rDNA Taxonomic Assignment
Phylogenetically classifies DNA sequence data using the nucleotide database and Taxonomy database from NCBI. The java-based software blasts each sequence that you provide and looks for a consensus taxon among the top-hits returned from blast. The software suite is published in Horton et al. 2010..
Effectorsearch
Quickly and effectively identify candidate type III secretion system effectors in bacterial genomes. Please reach out if you would like more information. The compressed archive file contains all scripts and dependencies as well as the text file Manual with instructions on installation and the contact details of developer Wen Zhang.
Arabidopsis thaliana Genetic Resources
Genomic Polymorphism Data
We have devoted substantial effort over the last decade establishing the ability to perform Genome Wide Association mapping in A. thaliana. This began with early work to understand genetic variation in the species, followed by later work to characterize patterns of linkage disequilibrium and the ability to association map simple traits. The work has come to fruition in a collaborative effort between the Bergelson, Borevitz and Nordborg labs in which 107 phenotypes are mapped (Atwell et al., 2010), and the associated database can be found on the GMI website.
Regional Mapping (RegMap) Lines
1,307 accessions of A. thaliana have been genotyped using the Affymetrix Arabidopsis 250k – SNP chip (Horton et al., 2012), after first selecting unique lines from a collection of ~6,000 accessions genotyped using 149 genome-wide SNPs (Platt et al., 2010). We have collected high quality geographic coordinates for 1,193 of these samples (Anastasio et al., 2011). Due to the extensive size of this collection, these accessions enable mapping using either global or regional populations, and thus allow to compare the genetic basis of ecologically important traits among distinct subsets (Horton et al., 2012). These genotyped lines were created in collaboration with the Borevitz and Nordborg laboratories, and are available through the Ohio State Arabidopsis Stock Center. SNPs genotypes are available here (note the file is a bit large: 1.1 GB). The latitude and longitude for these accessions are included in the archive. The annotations of individual SNPs are here. The regional mapping panel was used to search for candidate targets of selection using the pairwise haplotype sharing statistic (Toomajian et al., 2006), the composite likelihood ratio test (Nielsen et al., 2005), and a global FST scan (Lewontin & Krakauer, 1973). You can download these scores here. The map of recombination hot spots can be dowloaded here.
Climate Genome Scans
The Regmap lines were used to identify the genetics underlying climate adaptation in A. thaliana (Hancock et al., 2011). The archive includes a list of the top SNPs identified in this study. The climate data for 948 accessions is available here and explanation of the variables can be downloaded here. Please note that the genome browser allowing to view the results in their genomic context was taken offline.
1001 Genomes Project
Our group participates in the international collaboration that fully sequenced the genomes of 1001 A. thaliana accessions. More information can be found on the project website and the publication.
149-SNP Primers & Contaminant Identification
A set of primers for 149 SNPs has been established to quickly and cheaply fingerprint accessions, which is especially important in confirming the identify of stock center lines. Primer sequences can be downloaded here. We identified a set of Arabidopsis thaliana accessions that have previously been misidentified. The genetic fingerprint of 5965 accessions for the 149 SNPs and a list of putative contaminants among stock center lines can be downloaded here.
Various Lists of Accessions
Different studies have utilized specific sets of A. thaliana accessions:
- Members of the 196 mapping panel (e.g., Atwell et al., 2010)
- Justin Borevitz’s 360 lines
- Members of the 1307-RegMap panel
- Swedish accessions used in local adaptation studies